Like finally seeing all the gears of a watch and how they work together, researchers from UCLA and UC Berkeley have, for the first time ever, solved the puzzle of how the various components of an entire telomerase enzyme complex fit together and function in a three-dimensional structure.
The creation of the first complete visual map of the telomerase enzyme, which is known to play a significant role in aging and most cancers, represents a breakthrough that could open up a host of new approaches to fighting disease, the researchers said.
“Everyone in the field wants to know what telomerase looks like, and there it was. I was so excited, I could hardly breathe,” said Juli Feigon, a UCLA professor of chemistry and biochemistry and a senior author of the study. “We were the first to see it.”
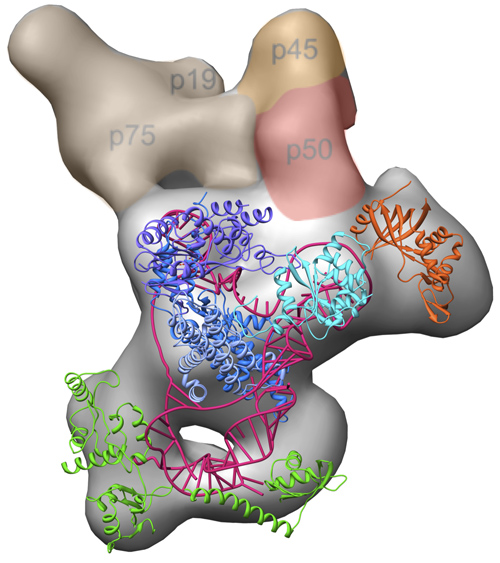
The three-dimensional electron microscopy structure of the complete Tetrahymena telomerase enzyme complex, with previously solved high-resolution structures modeled in. Image credit: Jiansen Jiang, Edward Miracco/UCLA Chemistry and Biochemistry
The scientists report the positions of each component of the enzyme relative to one another and the complete organization of the enzyme’s active site. In addition, they demonstrate how the different components contribute to the enzyme’s activity, uniquely correlating structure with biochemical function.
The research appears April 11 in the print edition of the journal Nature and is available online.
“We combined every single possible method we could get our hands on to solve this structure and used cutting-edge technological advances,” said co-first author Jiansen Jiang, a researcher who works with Feigon and the study’s co-senior author, Z. Hong Zhou, director of the Electron Imaging Center for Nanomachines at the California NanoSystems Institute at UCLA and a professor of microbiology, immunology and molecular genetics. “This breakthrough would not have been possible five years ago.”
“We really had to figure out how everything fit together, like a puzzle,” said co-first author Edward Miracco, a National Institutes of Health postdoctoral fellow in Feigon’s laboratory. “When we started fitting in the high-resolution structures to the blob that emerged from electron microscopy, we realized that everything was fitting in and made sense with decades of past biochemistry research. The project just blossomed, and the blob became a masterpiece.”
The telomerase enzyme is a mixture of components that unite inside our cells to maintain the protective regions at the ends of our chromosomes, which are called telomeres. Telomeres act like the plastic tips at the end of shoelaces, safeguarding important genetic information. But each time a cell divides, these telomeres shorten, like the slow-burning fuse of a time bomb. Eventually, the telomeres erode to a point that is no longer tolerable for cells, triggering the cell death that is a normal part of the aging process.
While most cells have relatively low levels of telomerase, 80 percent to 90 percent of cancer cells have abnormally high telomerase activity. This prevents telomeres from shortening and extends the life of these tumorigenic cells — a significant contributor to cancer progression.
The new discovery creates tremendous potential for pharmaceutical development that takes into account the way a drug and target molecule might interact, given the shape and chemistry of each component. Until now, designing a cancer-fighting drug that targeted telomerase was much like shooting an arrow to hit a bulls-eye while wearing a blindfold. With this complete visual map, the researchers are starting to remove that blindfold.
“Inhibiting telomerase won’t hurt most healthy cells but is predicted to slow down the progression of a broad range of cancers,” said Miracco. “Our structure can be used to guide targeted drug development to inhibit telomerase, and the model system we used may also be useful to screen candidate drugs for cancer therapy.”
The researchers solved the structure of telomerase in Tetrahymena thermophila, the single-celled eukaryotic organism in which scientists first identified telomerase and telomeres, leading to the 2009 Nobel Prize in medicine or physiology. Research on Tetrahymena telomerase in the lab of co-senior author Kathleen Collins, a professor of molecular and cell biology at UC Berkeley, laid the genetic and biochemical groundwork for the structure to be solved.
“The success of this project was absolutely dependent on the collaboration among our research groups,” said Feigon.
“At every step of this project, there were difficulties,” she added. “We had so many technical hurdles to overcome, both in the electron microscopy and the biochemistry. Pretty much every problem we could have, we had, and yet at each stage these hurdles were overcome in an innovative way.”
One of the biggest surprises, the researchers said, was the role of the protein p50, which acts as a hinge in Tetrahymena telomerase to allow dynamic movement within the complex; p50 was found to be an essential player in the enzyme’s activity and in the recruitment of other proteins to join the complex.
“The beauty of this structure is that it opens up a whole new world of questions for us to answer,” Feigon said. “The exact mechanism of how this complex interacts with the telomere is an active area of future research.”
“The atmosphere and collaboration at UCLA really amazes me, and that is combined with some of the most advanced facilities around,” Zhou said. “We have a highly advanced electron microscopy facility here at UCLA that even researchers without a strong background in electron microscopy can learn how to use and benefit from. This will be really useful as we move forward.”
Feigon’s research was federally funded by the National Science Foundation (grant MCB1022379) and the National Institutes of Health (grant GM48123). Kathleen Collins’ UC Berkeley research was funded by the NIH (grant GM54198); the scientists acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by the NIH (grant 1S10RR23057) to Zhou.
Equal contributions to the publication were made by co-first authors Jiang and Miracco, postdoctoral researchers at UCLA with Zhou and Feigon. Members of Kathleen Collins’ UC Berkeley laboratory who contributed to this research included technician Kyungah Hong, postdoctoral researcher Barbara Eckert and former graduate researcher Bosun Min. Other co-authors included Henry Chan and Darian D. Cash, UCLA graduate student researchers in Feigon’s laboratory.
– By Melody Pupols
*Source: University of California

