Enzymes evolved in the lab hold commercial and scientific promise
Whether big, small, slimy, or tall, most animal bodies are symmetric.
Except for sea anemones, starfish, sponges, and the like, animals have bilateral, or right-left, symmetry. Us included.
The bilateral body plan became the norm over eons of evolution. But what about molecules? Have any evolved common structures like a body plan?
Yes: the bundles of protein called enzymes, which carry out a sweeping variety of chemical reactions in living organisms. They have common features that also evolved over eons, but no fossil enzymes exist to reveal any ancestral versions.

Burckhard Seelig led a team that evolved a novel enzyme in a test tube. Image credit: University of Minnesota
Too bad, since new jobs for enzymes have arisen—like breaking down pollutants and making biofuels or drugs. If only we could, as in “Jurassic Park,” resurrect some of Nature’s discards and let them try their hand.
Enter University of Minnesota biochemist Burckhard Seelig. In earlier work as a postdoctoral fellow at Harvard, he evolved an enzyme that performs a useful function—joining pieces of RNA together—and has a unique structure that may be a throwback to earlier enzymes.
Using a modified version of PCR, the standard technique to generate multiple copies of genes, he and his colleagues generated trillions of DNA sequences, which they eventually turned into trillions of proteins, introducing mutations with each iteration of the process. They then screened each generation of proteins for any that could splice RNA and selected the one that performed best for further study. Now an assistant professor in the University’s Biotechnology Institute and Department of Biochemistry, Molecular Biology & Biophysics, Seelig, along with colleagues, has recently published a description of the enzyme’s structure in the journal Nature Chemical Biology.
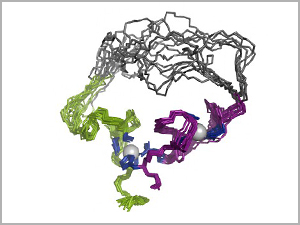
3-D structure of the evolved enzyme (an RNA ligase), using 10 overlaid snapshots. In the top region, the overlays show the range of bending and folding flexibility in the amino acid chain that forms the molecule. The two gray balls are zinc ions. Image credit: University of Minnesota
“It’s the first artificial enzyme evolved in a test tube,” says Seelig. “A handful of people worldwide make artificial enzymes, but they do it by rational design. They tinker with ones from nature that are already based on billions of years of evolution. But our enzyme is the product of just a few weeks of evolution.”
Seelig gives much of the credit for the research to graduate student Fa-An Chao, whom Seelig co-advises with his colleague Gianluigi Veglia.
Primordial protein?
The enzyme is a true original, a quality that has attracted not only commercial interests but NASA, which is one source of Seelig’s support.
“I see our work as a way to study the early history of functional proteins—that is, early life on Earth or life as one may find on other planets,” he says. “But most importantly, it’s a great way to tailor-make enzymes for different uses.”
Except with this method, enzyme designers won’t be bound by previous notions about how enzymes should look.
Newfangled fold
All proteins are composed of chains of amino acids. The “body plans,” or structures, of proteins, including enzymes, are achieved when the chains fold into the correct 3-D architectures, incorporating two basic features:
1) Twisted chains of amino acids called alpha helices (helixes); and/or
2) Flat chains of amino acids folded into hairpin curves like a paper clip (beta strands).
Both alpha helices and beta strands can stack up into bigger structures to increase the diversity of enzyme shapes, sizes, and functions. But Seelig’s new enzyme has neither alpha helices nor beta strands. Its folding is simpler, and two zinc ions act like chemical pins to hold the molecule together. He describes it as “small, wobbly, and flexible.”
But it does the job.
Seelig foresees a use for his enzyme in sequencing RNA, an essential step in deciphering the functions of numerous stretches of human DNA that were once thought useless and labeled “junk DNA.” It’s now known that those sequences contain master switches that control other genes and may hold the keys to understanding hundreds of baffling diseases, including autoimmune disorders.
*Source: University of Minnesota
