University of Arizona researcher Michael Worobey and his team have discovered that the key to understanding influenza pandemics may lie in flu exposure during childhood
Just as the world was recovering from the devastation of World War I, another killer swept across the globe. A deadly flu virus attacked more than one-third of the world’s population, and within months had killed more than 50 million people – three times as many as the war – and had done it more quickly than any other illness in recorded history.
Until now, the origin of the 1918 pandemic flu virus and its unusual severity have vexed health experts. A new study led by Michael Worobey, professor in the University of Arizona Department of Ecology and Evolutionary Biology, within of the College of Science, not only sheds light on the devastating 1918 pandemic, but could also improve vaccination strategies, and pandemic prevention and preparedness.
“If our model is correct, then current medical interventions, especially antibiotics and vaccines, against several pneumonia-causing bacteria, could be expected to dramatically reduce mortality, if we were faced today with a similar set of pandemic ingredients,” Worobey said.
“Ever since the great flu pandemic of 1918, it has been a mystery where that virus came from and why it was so severe and, in particular, why it killed young adults in the prime of life,” Worobey added. “It has been a huge question whether there was something special about that situation, and whether we should expect the same thing to happen tomorrow.”
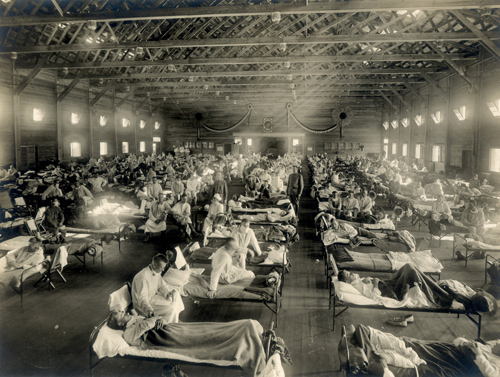
An emergency hospital in Camp Funston, Kansas, during the 1918 influenza epidemic. (Photo credit: Otis Historical Archives, National Museum of Health & Medicine)
Worobey and his colleagues developed an unprecedentedly accurate “molecular clock” approach to untangle the origins of the 1918 pandemic H1N1 influenza A virus, the classical swine H1N1 influenza virus, and the post-pandemic seasonal H1N1 lineage that circulated from 1918 until 1957. A molecular clock is a technique used in evolutionary biology to reconstruct family trees of organisms – or viruses – based on the number of genetic mutations accumulating over time.
The researchers discovered that the pandemic virus arose shortly before 1918, when a human H1 virus, which they infer had already been circulating in the human population since about 1900, picked up genetic material from a bird flu virus.
The Human influenza A virus usually sees higher mortality rates for infants and the elderly, but the pandemic virus caused extensive deaths in people ages 20 to 40, primarily from secondary bacterial infections, especially pneumonia.

Michael Worobey is a professor in the UA Department of Ecology and Evolutionary Biology. In previous research projects, he traced the origins of HIV in Africa and how the virus spread across the world. (Photo credit: Beatriz Verdugo/UANews)
The authors suggest that this likely was because many young adults born from about 1880 to 1900 were exposed during childhood to a putative H3N8 virus circulating in the population, which had surface proteins that were very different from those of the H1N1 virus. In contrast, most individuals born earlier or later than 1880-1900 would have had better protection because they were more likely to have been exposed a virus variant more similar to the 1918 virus.
“You can picture the flu virus as a tiny soccer ball studded with lollipops,” Worobey explained. “The candy part of the lollipop is by far the most potent part of the flu virus, against which our immune system can make antibodies. If antibodies cover all the lollipop heads, the virus can’t even infect you.”
For example, a person exposed to a flu strain studded with blue candy lollipops would have antibodies protecting it from future infections with that flu virus. However, if faced with a different strain containing red candy, that same person would get sick because the “anti-blue” antibodies would not recognize the red lollipops, allowing the virus to slip through the body’s immune defenses unscathed.
“We believe that the mismatch between antibodies trained to H3 virus protein and the H1 protein of the 1918 virus may have resulted in the heightened mortality in the age group that happened to be in their late 20s during the pandemic,” Worobey said.
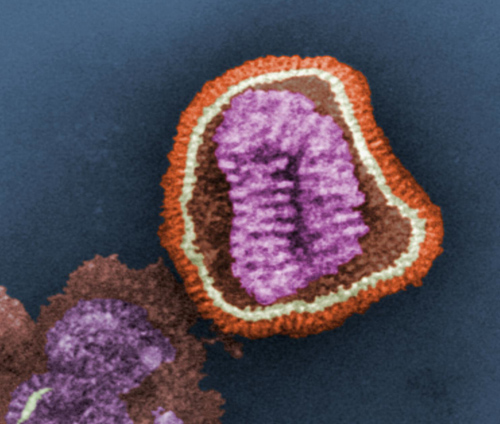
A false-color image of influenza virus as seen through an electron microscope. The virus’ genetic material (purple) is encased in a shell with surface proteins (red) used to infect a host cell. (Photo credit: U.S. Centers for Disease Control Public Library)
Worobey said the new perspective applies not only to the pandemic of 1918, but might also explain patterns of seasonal flu mortality and the mysterious patterns of mortality when humans are infected by highly pathogenic avian-origin H5N1 and H7N9 viruses, commonly known as the “bird flu.”
H5N1causes higher mortality rates in young people, and H7N9 causes higher mortality in the elderly. In both cases, the researchers found that the more susceptible age groups were exposed initially, as children, to viruses with a mismatched HA, and may suffer severe consequences similar to young adults faced with a mismatched virus in 1918.
The authors suggest that immunization strategies mimicking the often impressive protection provided by initial childhood exposure to influenza virus variants encountered later in life might dramatically reduce mortality due to both seasonal and novel IAV strains.
Earlier this year, Worobey and his collaborators published a study in the journal Nature providing the most comprehensive analysis to date of the evolutionary relationships of flu virus across different host species over time. The results revealed a rapid, global replacement of the genes in the avian flu virus coinciding closely with a horse flu outbreak in the 1870s that crippled the economy. They also challenged the accepted wisdom of wild birds as the major reservoir harboring the flu virus, from where it jumps to domestic birds and other species, including humans. Instead, the researchers found a very strong indication of spillover from domestic birds to wild birds.
The latest study is published in the early edition of the Proceedings of the National Academy of Sciences. It is co-authored by Guan-Zhu Han at the UA Department of Ecology and Evolutionary Biology and Andrew Rambaut at the Centre for Infection, Immunity and Evolution at the University of Edinburgh, and the National Institutes of Health in Bethesda, Maryland.
– By Daniel Stolte
*Source: The University of Arizona
