*Looking in vivo at virus-bacterium associations sets stage for better understanding of such interactions in human health*
Viruses are the most abundant parasites on Earth. Well known viruses, such as the flu virus, attack human hosts, while viruses such as the tobacco mosaic virus infect plant hosts.
More common, but less understood, are cases of viruses infecting bacteria known as bacteriophages, or phages. In part, this is due to the difficulty of culturing bacteria and viruses that have been cut off from their usual biological surroundings in a process called in vitro.
Researchers from the California Institute of Technology, funded in part by the National Science Foundation, were the first to use a clever technique to look at virus-bacterium interactions in vivo, that is, within an organism’s normal state. The researchers report their results in the July 1st issue of the journal, Science.
As a test case, Rob Phillips and his team considered the interaction between viruses and bacteria in the hindgut, or posterior part, of a termite. Using new microfluidic technology, they were able to isolate single bacterial cells from the termite hindgut in six-nanoliter chambers on an array containing 765 such chambers.
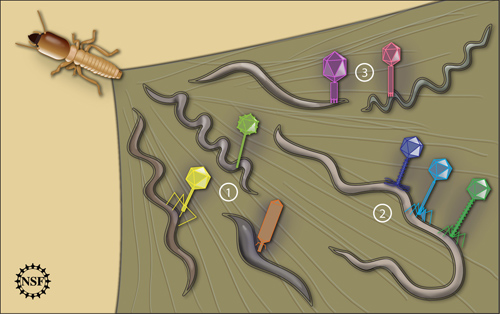
Virus-bacterium associations were examined in the natural environment of a termite's hindgut. Three general scenarios were seen. In the first (1) there was a one-to-one association: one type of virus matched one type of bacterium host. In the second (2) the host bacterium was associated with a diverse group of viruses, indicating perhaps a more ancient infection or a more susceptible host. In the third case (3) very similar viruses were seen infecting several different types of bacterial hosts. This study tested methods of examining virus-bacterium interactions in nature, rather than in vitro--from a culture. It opens a new door to understanding the diverse and highly populated world of viruses and bacteria that we know so little about. Image credit: Zina Deretsky, National Science Foundation
They were then able to determine whether the chambers contained bacterial DNA, viral DNA or both. In the latter case, the researchers were able to statistically deduce whether the virus was specifically associated with the host–for example, by attaching to the host, shooting its DNA into the host, being incorporated into the host as a prophage (a viral genome inserted and integrated into the bacterial DNA), riding on a plasmid or by assembling new viruses within the host. And through this snapshot, the group recorded virus-bacterium associations.
Frequently there was a one-to-one virus-bacterium correspondence. However in some cases, the host was associated with a viral gene exhibiting marked diversity, suggesting possibly a more ancient infection, a more susceptible host or a phage replicating at a lower fidelity. By analyzing the bacteria and viruses based on their evolutionary development they were able to deduce that horizontal gene transfer, while it may be occurring, is not occurring at a rate high enough to randomize host-virus associations.
This study was by no means exhaustive. Many similar associations may still be found in the termite hindgut. And further inquiry may lead to a better understanding of the coevolution of a virus and its host. However, this was the first in vivo exercise, and it opens the doors of the field much wider than previously possible through in vitro culture alone.
*Source: National Science Foundation (NSF)
