Berkeley Lab Finding that Protein Folding Funnels Also Apply to Self-Assembly Should Benefit Biomimicry and Nanosynthesis
Proteins are able to self-assemble into a wide range of highly ordered structures that feature a diverse array of properties. Through biomimicry – technological innovation inspired by nature – humans hope to emulate proteins and produce our own version of self-assembling molecules. A key to accomplishing this is understanding how protein-folding – a process critical to the form and function of a protein – is extended from individual proteins to complex assemblies.
Researchers with the U.S. Department of Energy (DOE)’s Lawrence Berkeley National Laboratory (Berkeley Lab) have now shown that a concept widely accepted as describing the folding of a single individual protein is also applicable to the self-assembly of multiple proteins. Their findings provide important guidelines for future biomimicry efforts, particularly for device fabrication and nanoscale synthesis.
“We’ve made the first direct observations that the concept of a folding funnel with kinetic energy traps for individual proteins can be equally applied to the assembly of ordered protein structures,” says Jim DeYoreo, a scientist with the Molecular Foundry, a DOE nanoscience center at Berkeley Lab, who led this research along with Berkeley Lab chemist Carolyn Bertozzi. “Our results tell us that efforts to discover and codify the design rules for the self-assembly of complex molecular systems will have to take into account the impact of kinetic traps associated with conformational transformations.”
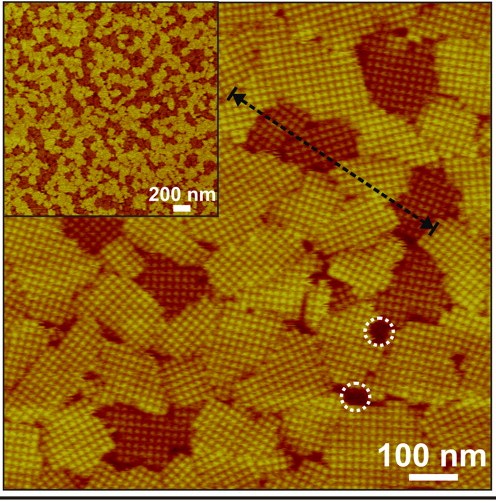
AFM micrograph of 2D S-layers assembled on mica shows two different pathways to crystalization, one in which the domans are 2-3 nanometers taller (white circles). Height differences, measured along the dotted black line, were the result of kinetic trapping. (Image from Molecular Foundry)
DeYoreo and Bertozzi are the corresponding authors of a paper published by the Proceedings of the National Academy of Sciences (PNAS) that reported this research. The paper is titled “Direct observation of kinetic traps associated with structural transformations leading to multiple pathways of S-layer assembly.” Co-authoring the paper were Seong-Ho Shin, Sungwook Chung, Babak Sanii and Luis Comolli.
Proteins are essentially biomolecular nanomachines capable of performing numerous tasks because of their ability to fold themselves into a multitude of shapes and forms. When individual proteins self-assemble into ordered structures the resulting ensemble often adopts conformations that are quite distinct from those of the individual components.
“For example, collagen matrices, which constitute the organic scaffolds of bones and teeth, are constructed from triple helices of individual collagen monomers,” DeYoreo says. “These helices will further assemble into highly organized twisted fibrils that exhibit a pseudohexagonal symmetry.”
The folding funnel concept explains individual protein folding on the basis of conformational changes to reach a state of minimal free energy. An unfolded protein starts out in a state of high free energy that makes its conformation unstable. Initially, there are a number of possible three-dimensional conformations that would reduce this free energy. However, as the protein starts to fold, the free energy begins to drop and the number of possible conformations begins to decrease like the shrinking width of a funnel. The bottom of the funnel is reached when free energy is minimized and there is only one available conformation. As the free energy drops, however, there may be kinetic traps along the way that can stop the folding process and hold the protein in partially folded conformations, known as molten globules and folding intermediates, for extended periods of time. Eventually these trapped conformational states will be transformed into a stable conformation but the shape and form of that final conformation is influenced by the kinetic traps.
“In a protein folding funnel, the funnel walls are presumed not to be smooth and the resulting bumps and valleys define kinetic traps,” DeYoreo says. “This physical picture of folding has been explored in some detail at the single molecule level, but has not been considered for protein self-assembly into extended architectures even though conformational transformations are part and parcel of the self-assembly process.”
(From left) Sungwook Chung, Seong-Ho Shin, James DeYoreo and Carolyn Bertozzi with Berkeley Lab’s Molecular Foundry, were part of a team that demonstrated the concept of folding funnels applies equally to individual and ensembles of proteins. (Photo by Roy Kaltschmidt)
DeYoreo, Bertozzi and their colleagues took steps to correct this knowledge deficit by studying the surface-layer (S-layer) proteins that self-assemble into a crystalline membrane around the single cells of bacteria and Archaea. This outer membrane serves as the first point of contact between the microbe and its environment and is key to the microbe’s ability to survive. Using in situ Atomic Force Microscopy (AFM), the researchers imaged in real time and at the molecular level kinetic trapping during the 2D self-assembly of S-layer protein structures on mica surfaces.
“We observed that self-assembly of S-layer proteins tracks along two different pathways, one leading directly to the low-energy final, ordered state, and the other leading to a kinetic trap occupied by a long-lived transient state that is more disordered,” DeYoreo says. “Although either state is easily accessible during crystal nucleation, if the system falls into the high-energy state, escape to the final, low-energy state is strongly impeded at room temperature. This demonstrates the importance of kinetic traps in determining the pathway of S-layer crystallization and suggests that the concept of folding funnels is equally valid for self-assembly of extended protein structures.”
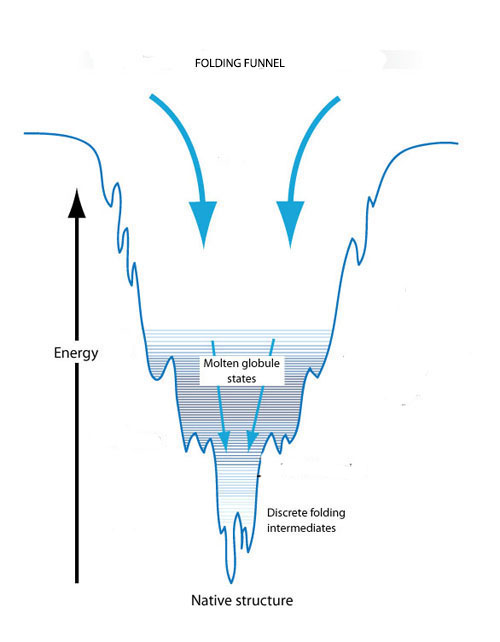
The folding funnel concept explains protein folding on the basis of conformational changes to reach a state of minimal free energy. An unfolded protein starts out in a state of high free and reaches its native state at minimal energy. Kinetic traps along the way can create transient molten globules and folding intermediates. Image credit: Berkeley Lab
This research was supported by the DOE Office of Science.
– By Lynn Yarris
*Source: Berkeley Lab
